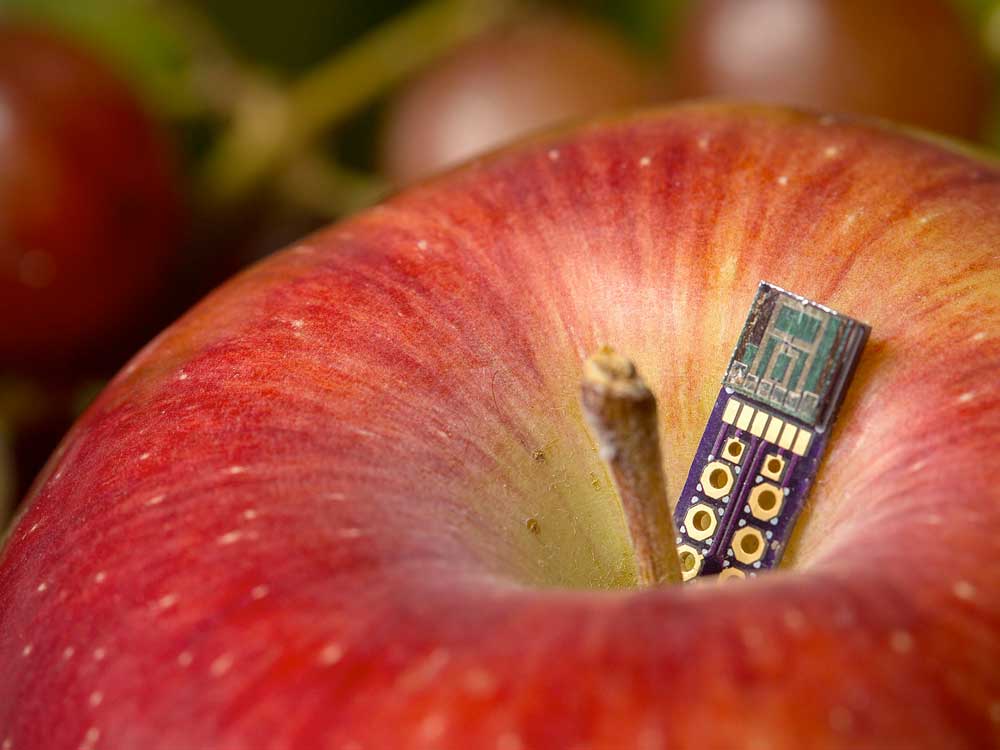
The way plants interact with their environments are shaped by their genetic makeup and the complex biological networks within them. These interactions determine how plants grow, adapt, and thrive across diverse conditions. By studying these relationships in depth, we can unlock new insights into how plants function and evolve.
Our research focuses on how genetics and environmental factors interact with programmed plants. By applying computational modeling and advanced measurement tools, the team aims to explore previously inaccessible traits and discover how plants adapt to changing environments. This work has the potential to expand the adaptive capacity of crops, improve sustainability, and enhance productivity across a wide range of agricultural systems.